Test-retest reproducibility¶
We first quantified the spatial reproducibility of parcels in friends-s01 training and validation data.
Reproducibility measure. A spatial correlation is generated for each pair of parcels, one in the training and one in the validation set. This spatial correlation is calculated within the union of the subject-specific grey matter mask from both parcellations.
Intra-subject reproducibility. For each parcel in the training set, we report the maximal spatial correlation with any parcels of the validation. This process generates a distribution of intra-subject reproducibility across parcels.
Inter-subject reproducibility. We also apply the same matching procedure, but comparing pairs of parcels coming from different subjects, within the training set. This generates a distribution of inter-subject reproducibility across parcels.
Computational reproducibility
The results for this experiment have been precomputed usin the scripts bla and bla. The following hidden block of code loads the necessary libraries to generate the figures, sets the path to find the pre-comupted results as well as some visualisation parameters.
import os
from cneuromod_embeddings import dypac_utils
from cneuromod_embeddings.repro_summary import repro_df
import seaborn as sns
import matplotlib.pyplot as plt
root_path, basename = os.path.split(dypac_utils.__file__)
root_data = os.path.join(root_path, 'friends-s01', 'repro_friends-s01')
val_repro = repro_df(root_data)
params = dypac_utils.dypac_params()
sns.set_theme(style="whitegrid")
sns.set(font_scale=1.5)
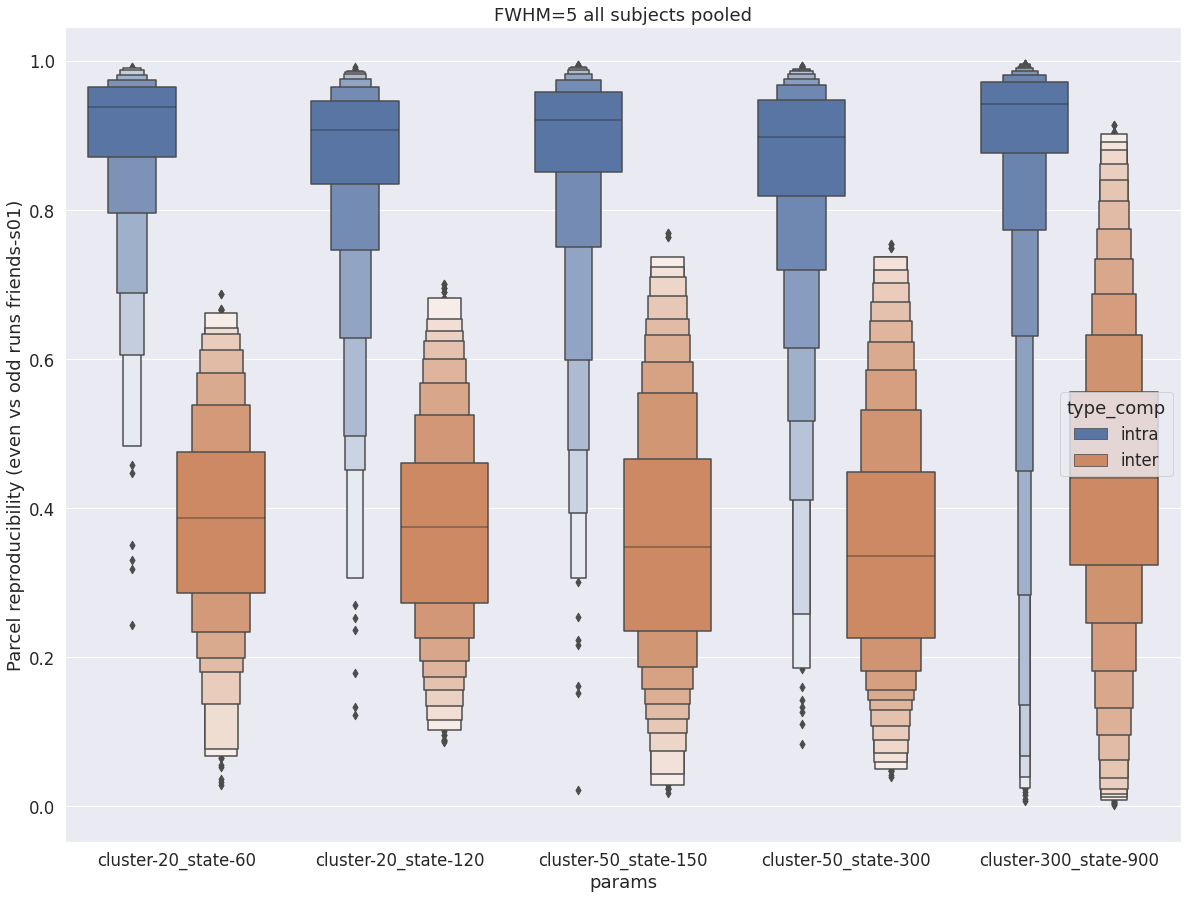
The following figure shows the distribution of intra- vs inter-subject parcel reproducibility for fwhm=5. The intra-subject reproducibility is very high for all paramaters, with median reproducibility above 0.9 in all cases. The inter-subject reproducibility is substantially lower than intra-subject reproducibility, with slightly higher median and variance for larger cluster and state. Overall, the largest (and near-perfect) separation intra- vs inter-subject is observed for cluster-20_state-60, indicating that the major states of large distributed networks are more subject-specific than local functional regions.
fwhm=5
fig = plt.figure(figsize=(20, 15))
data = val_repro[val_repro['fwhm']==fwhm]
sns.boxenplot(data=data, x='params', y='spatial_r', hue='type_comp', scale='area')
plt.ylabel('Parcel reproducibility (even vs odd runs friends-s01)')
plt.title(f'FWHM={fwhm} all subjects pooled')
Text(0.5, 1.0, 'FWHM=5 all subjects pooled')
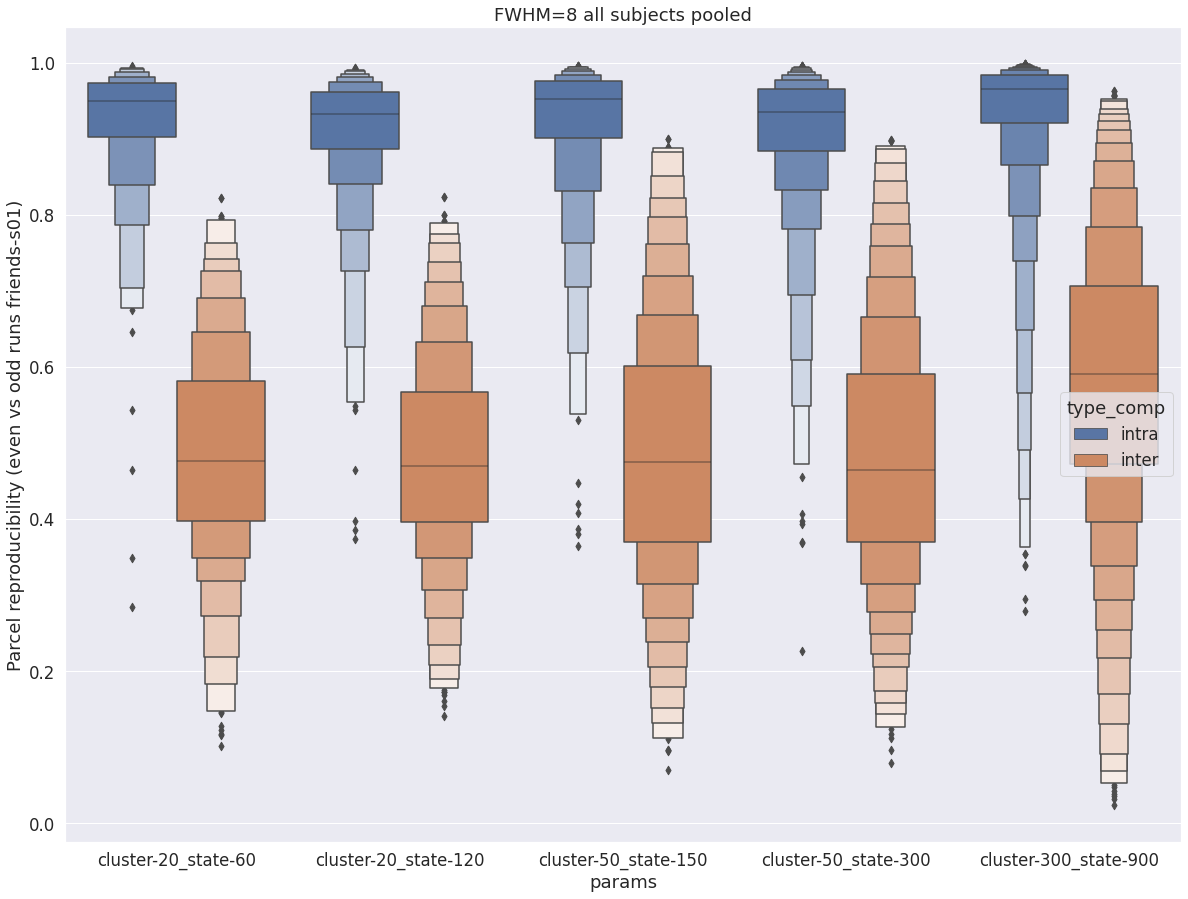
Repeating the experiment with fhwm=8 leads to similar conclusions, with two notable differences: (1) the distributions of intra-subject reproducibility gets tighter, (2) inter-subject reproducibility increases, and susbstantially overlaps intra-subject distribution for cluster-300_state-900. This suggests that higher levels of spatial blurring help stabilize the dynamic parcels, but also removes some subject-specific details.
fwhm=8
fig = plt.figure(figsize=(20, 15))
data = val_repro[val_repro['fwhm']==fwhm]
sns.boxenplot(data=data, x='params', y='spatial_r', hue='type_comp', scale='area')
plt.ylabel('Parcel reproducibility (even vs odd runs friends-s01)')
plt.title(f'FWHM={fwhm} all subjects pooled')
Text(0.5, 1.0, 'FWHM=8 all subjects pooled')
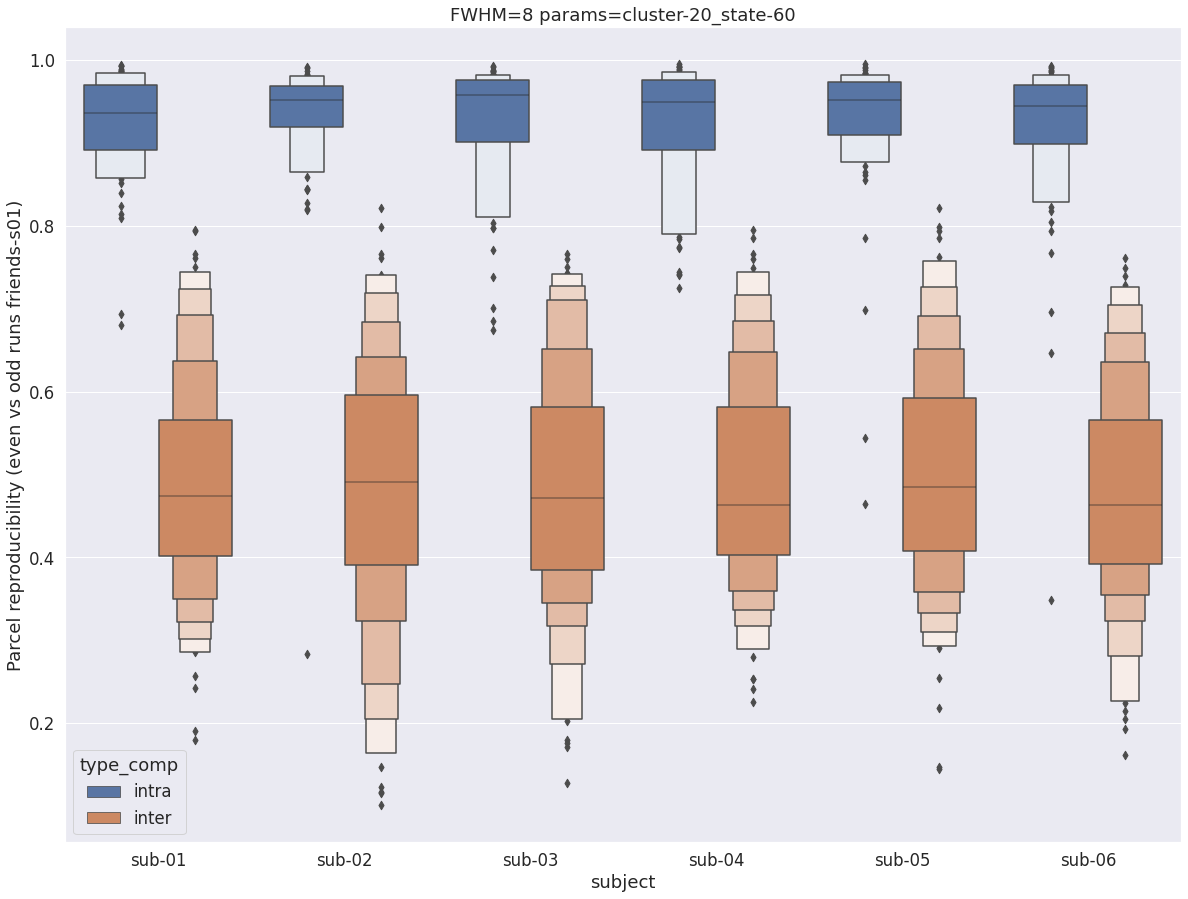
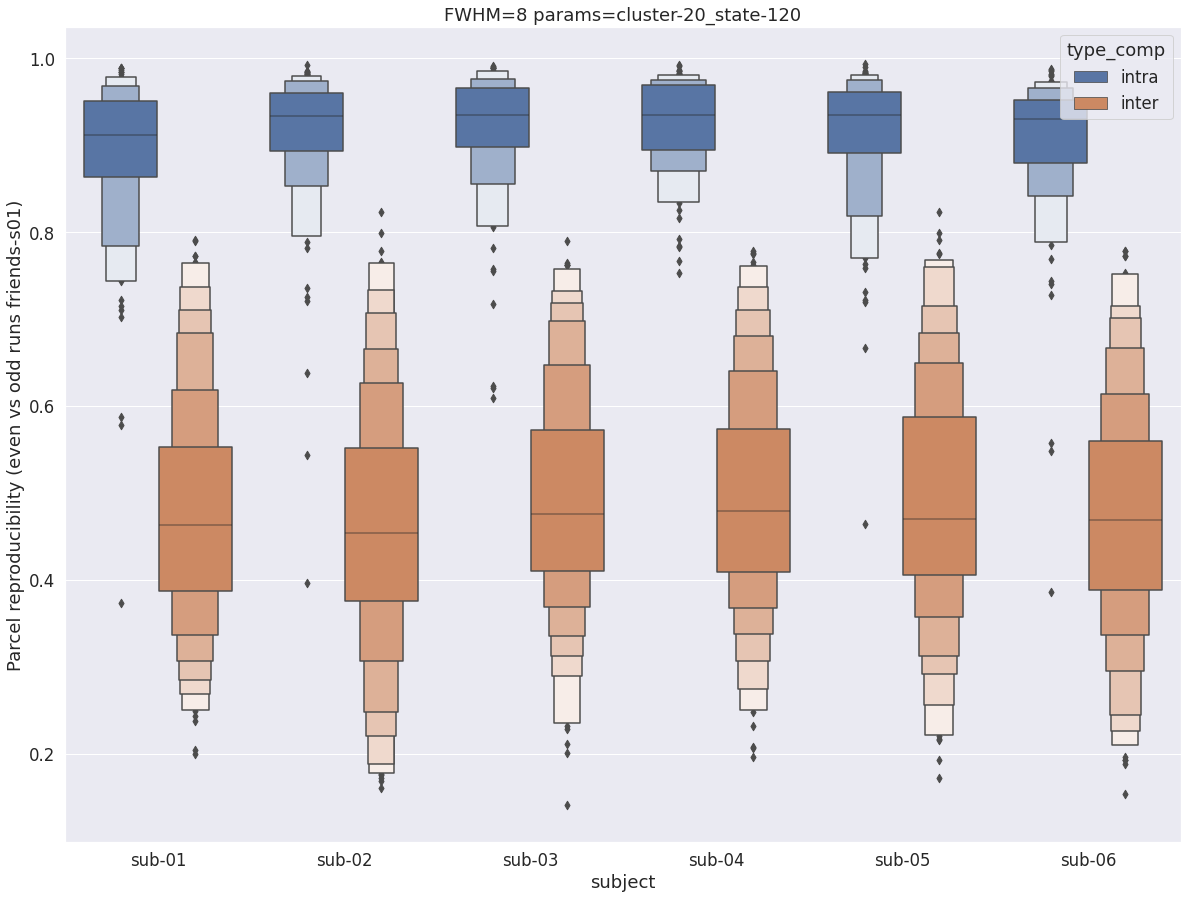
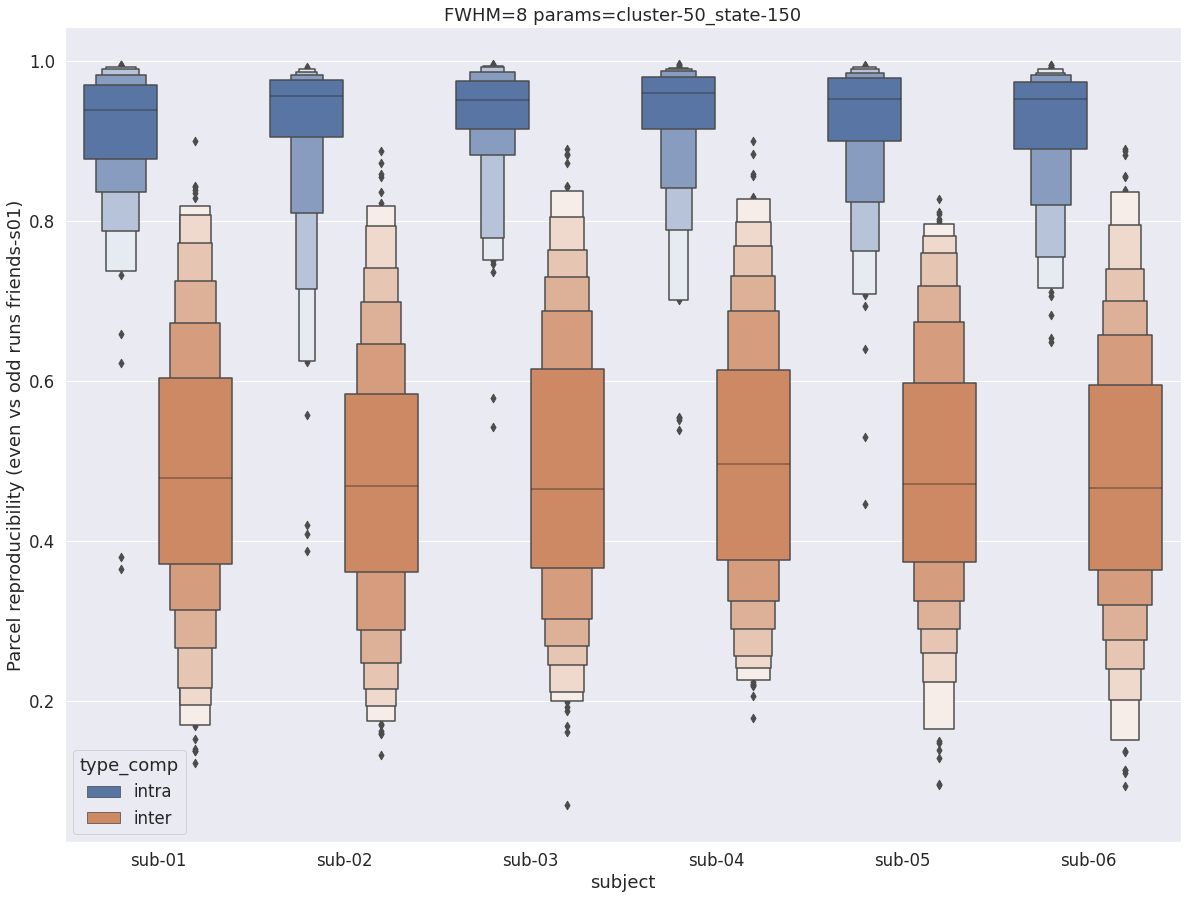
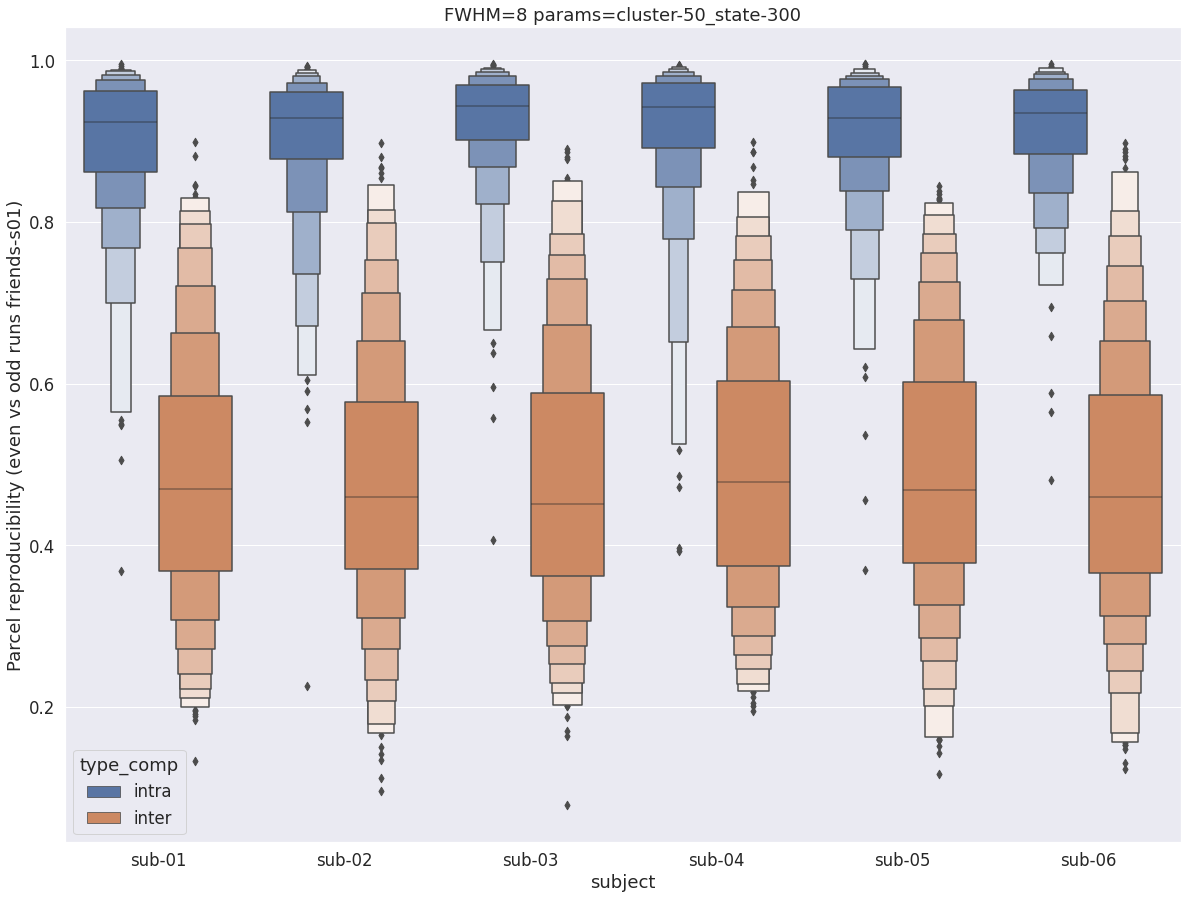
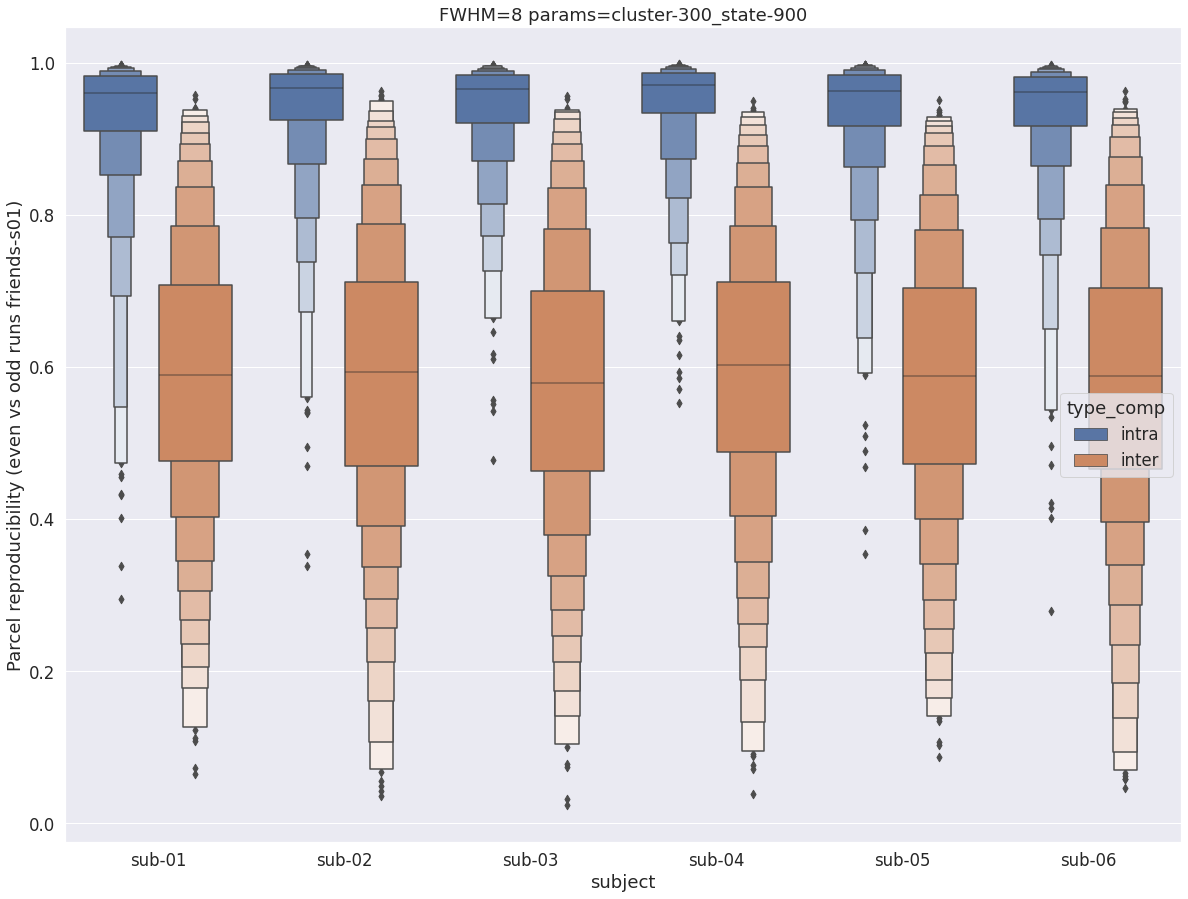
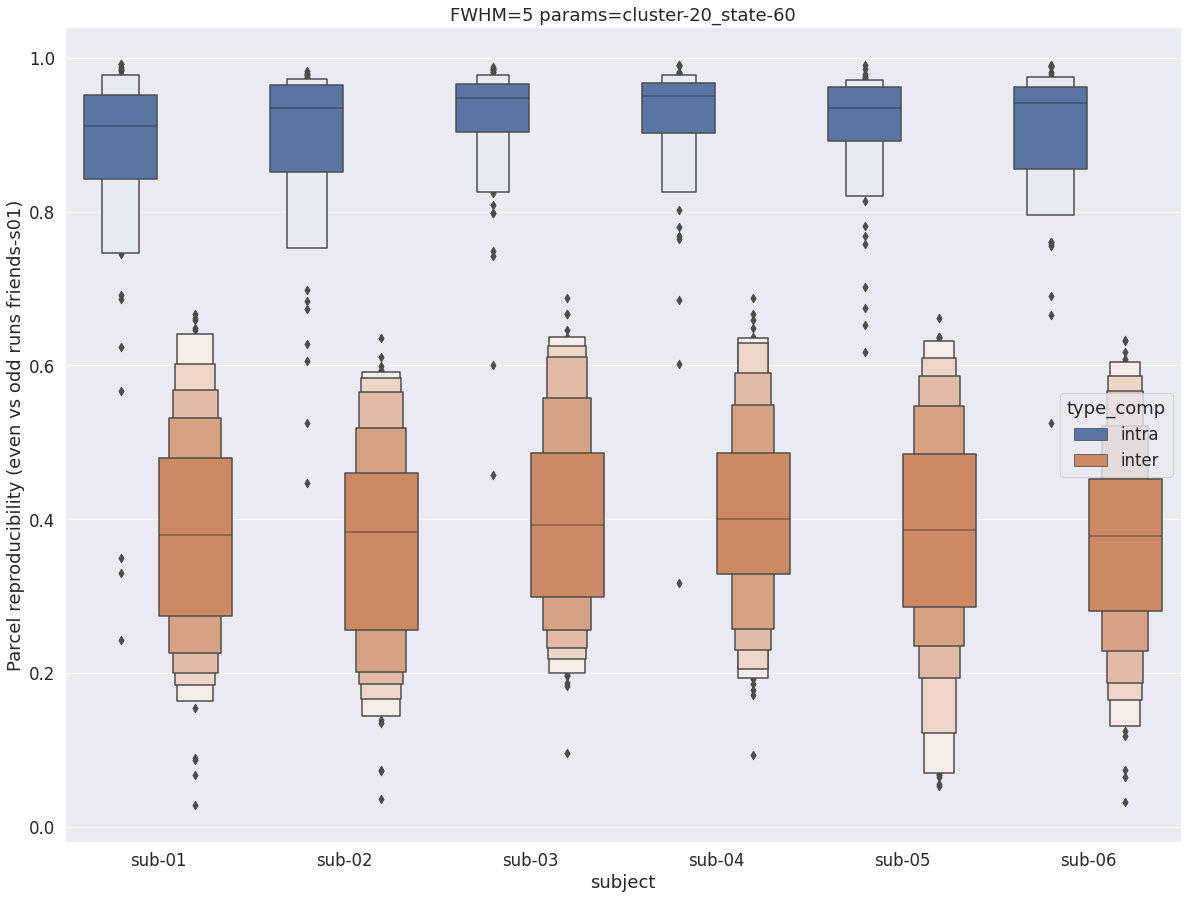
Finally, for a given set of parameters, it is possible to further break down the reproducibility distributions per subject, which is shown in the following supplementary figures. The intra- vs inter-subject distribution of spatial reproducibility is strikingly consistent across subjects, both for fwhm=5 and for fwhm=8.
fwhm=5
for ind, cluster in enumerate(params['cluster']):
state = params['state'][ind]
key = f'cluster-{cluster}_state-{state}'
fig = plt.figure(figsize=(20, 15))
data = val_repro[val_repro['fwhm']==fwhm][val_repro['params']==key]
sns.boxenplot(data=data, x='subject', y='spatial_r', hue='type_comp', scale='area')
plt.ylabel('Parcel reproducibility (even vs odd runs friends-s01)')
plt.title(f'FWHM={fwhm} params={key}')
/home/pbellec/env/cneuromod_embeddings/lib/python3.6/site-packages/ipykernel_launcher.py:6: UserWarning: Boolean Series key will be reindexed to match DataFrame index.
/home/pbellec/env/cneuromod_embeddings/lib/python3.6/site-packages/ipykernel_launcher.py:6: UserWarning: Boolean Series key will be reindexed to match DataFrame index.
/home/pbellec/env/cneuromod_embeddings/lib/python3.6/site-packages/ipykernel_launcher.py:6: UserWarning: Boolean Series key will be reindexed to match DataFrame index.
/home/pbellec/env/cneuromod_embeddings/lib/python3.6/site-packages/ipykernel_launcher.py:6: UserWarning: Boolean Series key will be reindexed to match DataFrame index.
/home/pbellec/env/cneuromod_embeddings/lib/python3.6/site-packages/ipykernel_launcher.py:6: UserWarning: Boolean Series key will be reindexed to match DataFrame index.
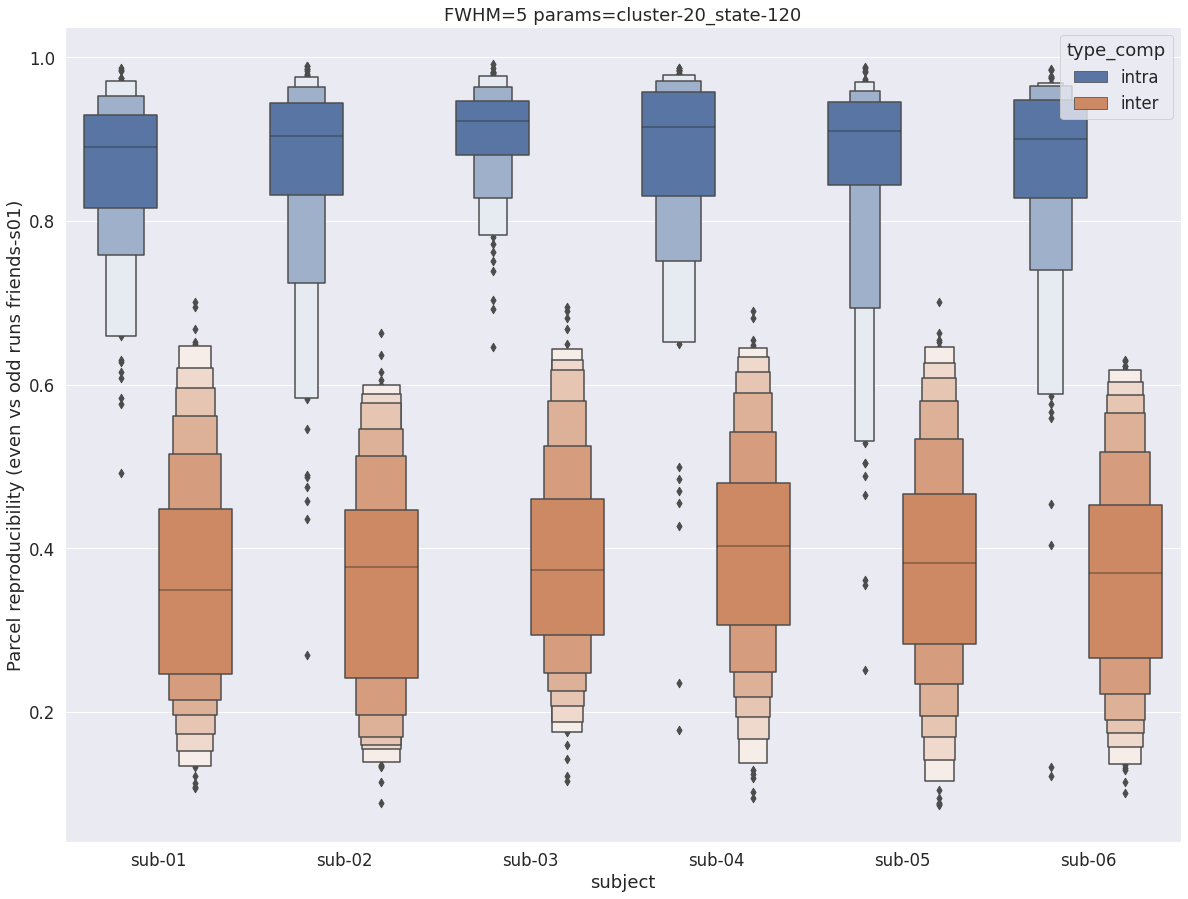
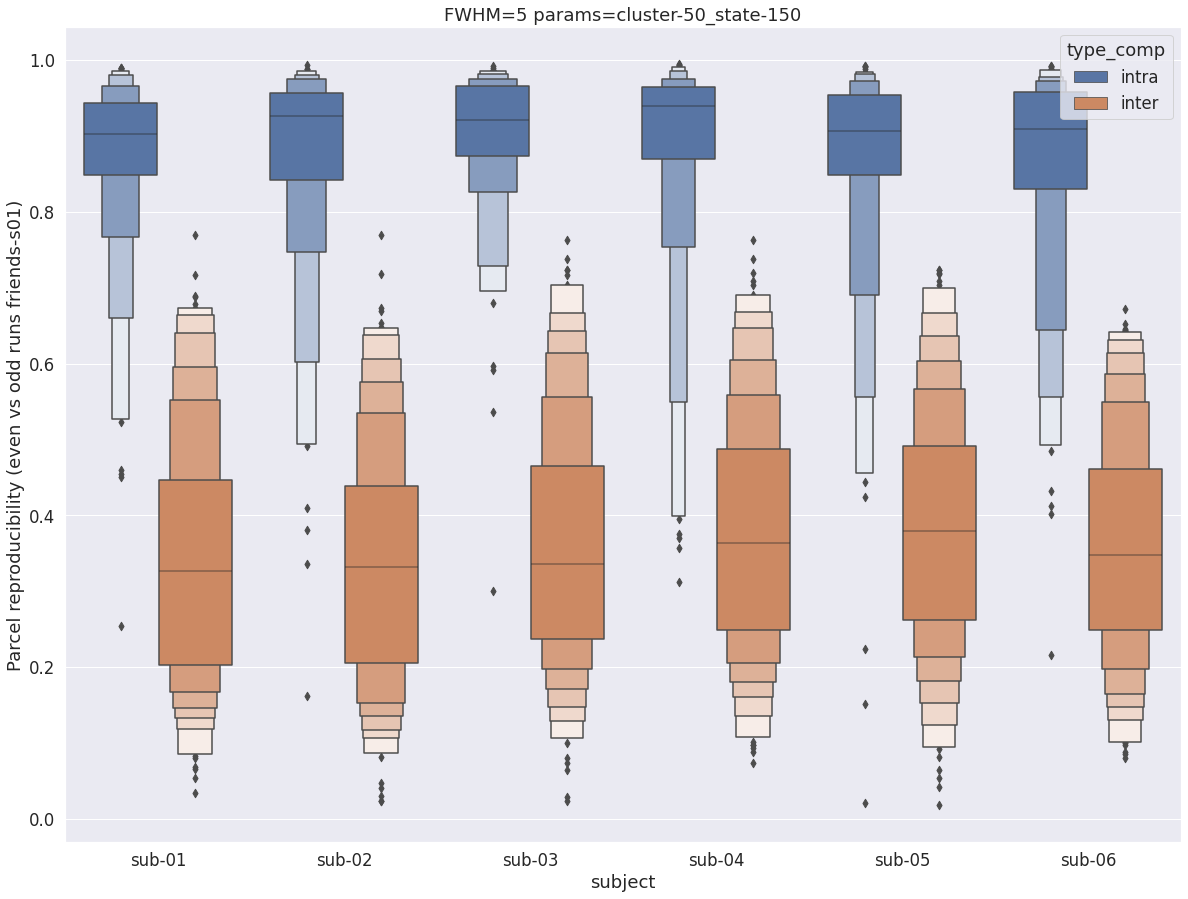
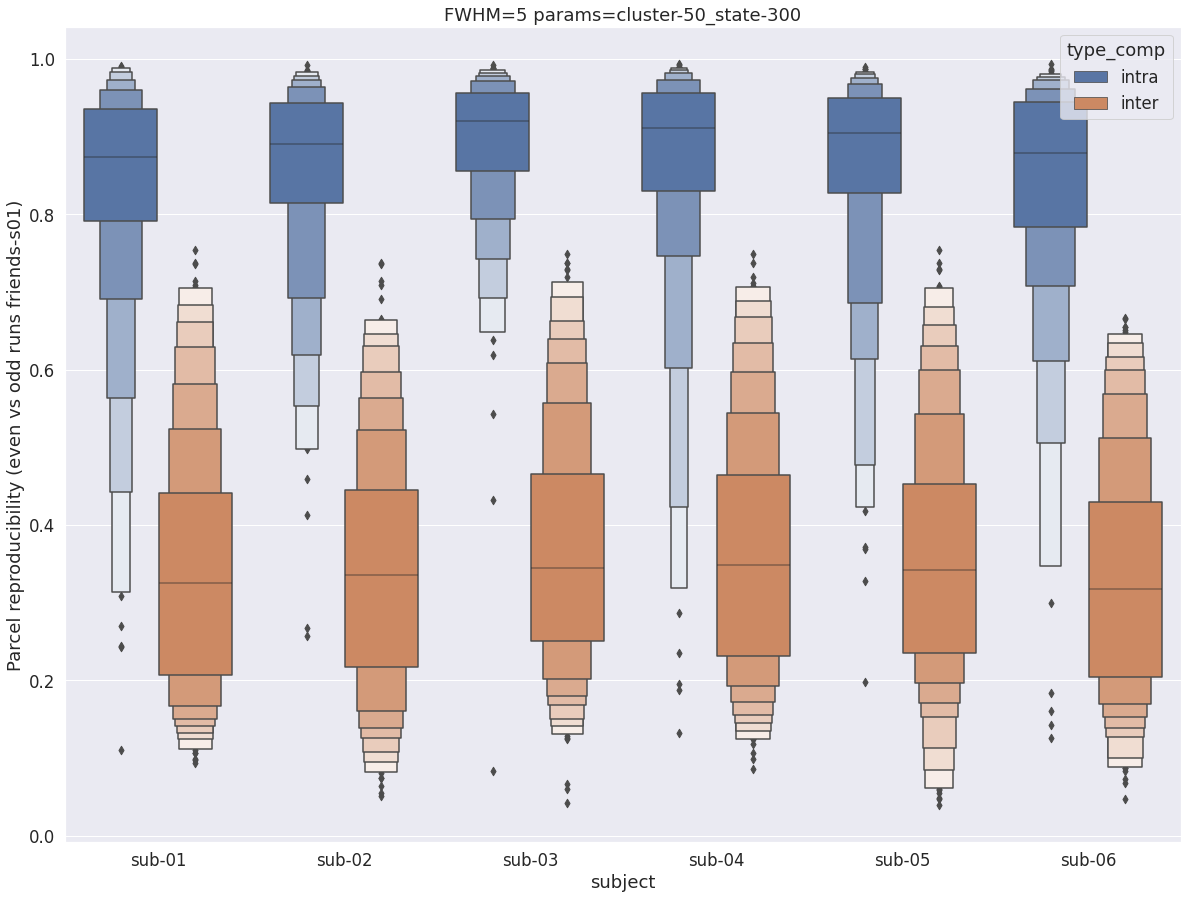
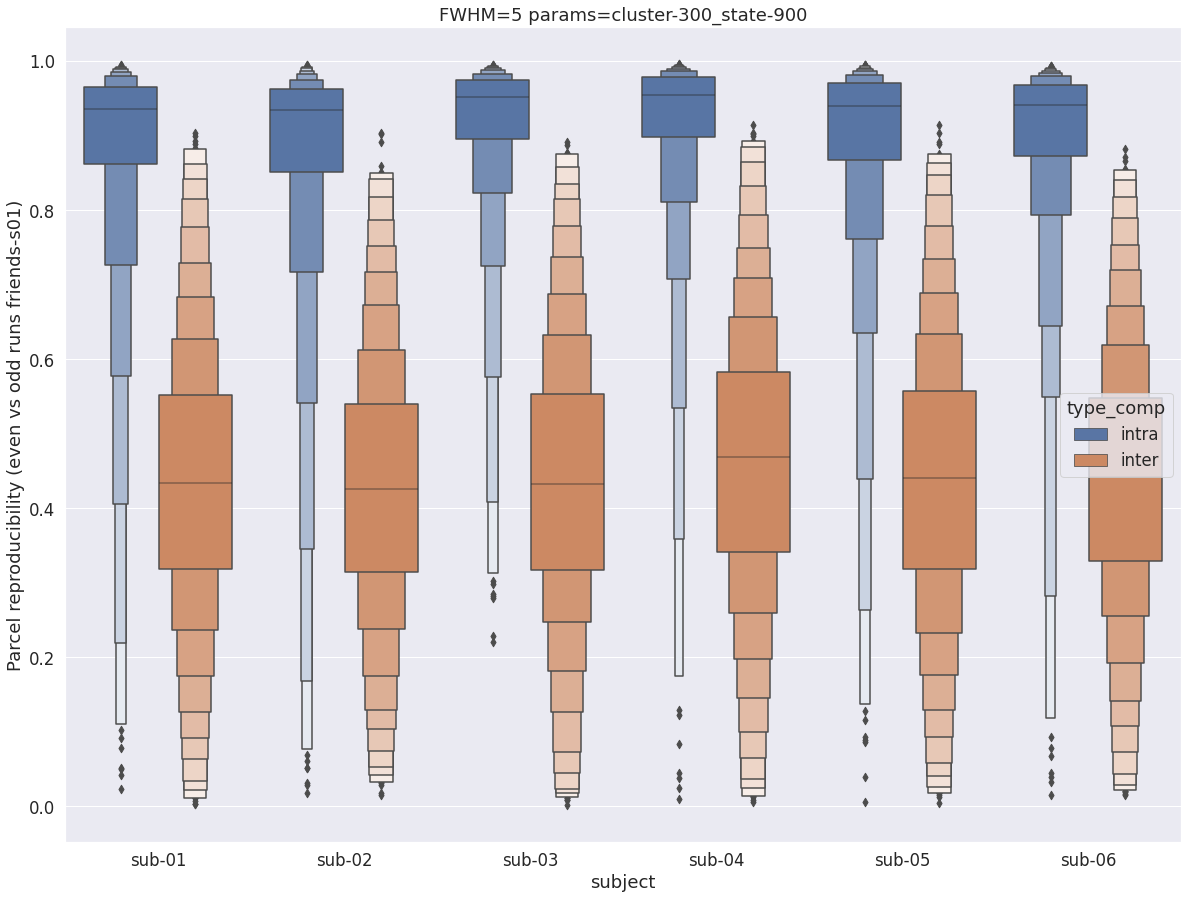
fwhm=8
for ind, cluster in enumerate(params['cluster']):
state = params['state'][ind]
key = f'cluster-{cluster}_state-{state}'
fig = plt.figure(figsize=(20, 15))
data = val_repro[val_repro['fwhm']==fwhm][val_repro['params']==key]
sns.boxenplot(data=data, x='subject', y='spatial_r', hue='type_comp', scale='area')
plt.ylabel('Parcel reproducibility (even vs odd runs friends-s01)')
plt.title(f'FWHM={fwhm} params={key}')
/home/pbellec/env/cneuromod_embeddings/lib/python3.6/site-packages/ipykernel_launcher.py:6: UserWarning: Boolean Series key will be reindexed to match DataFrame index.
/home/pbellec/env/cneuromod_embeddings/lib/python3.6/site-packages/ipykernel_launcher.py:6: UserWarning: Boolean Series key will be reindexed to match DataFrame index.
/home/pbellec/env/cneuromod_embeddings/lib/python3.6/site-packages/ipykernel_launcher.py:6: UserWarning: Boolean Series key will be reindexed to match DataFrame index.
/home/pbellec/env/cneuromod_embeddings/lib/python3.6/site-packages/ipykernel_launcher.py:6: UserWarning: Boolean Series key will be reindexed to match DataFrame index.
/home/pbellec/env/cneuromod_embeddings/lib/python3.6/site-packages/ipykernel_launcher.py:6: UserWarning: Boolean Series key will be reindexed to match DataFrame index.