R2 in the cerebellum¶
This analysis is supplementary to the results reported in R2 in the cortex. The difference is that here the summary statistics are computed in a mask including the cerebellum, instead of the cortex.
import os
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
from cneuromod_embeddings.r2_summary import _r2_intra, _r2_inter, _r2_other
sns.set_theme(style="whitegrid")
sns.set(font_scale=1.5)
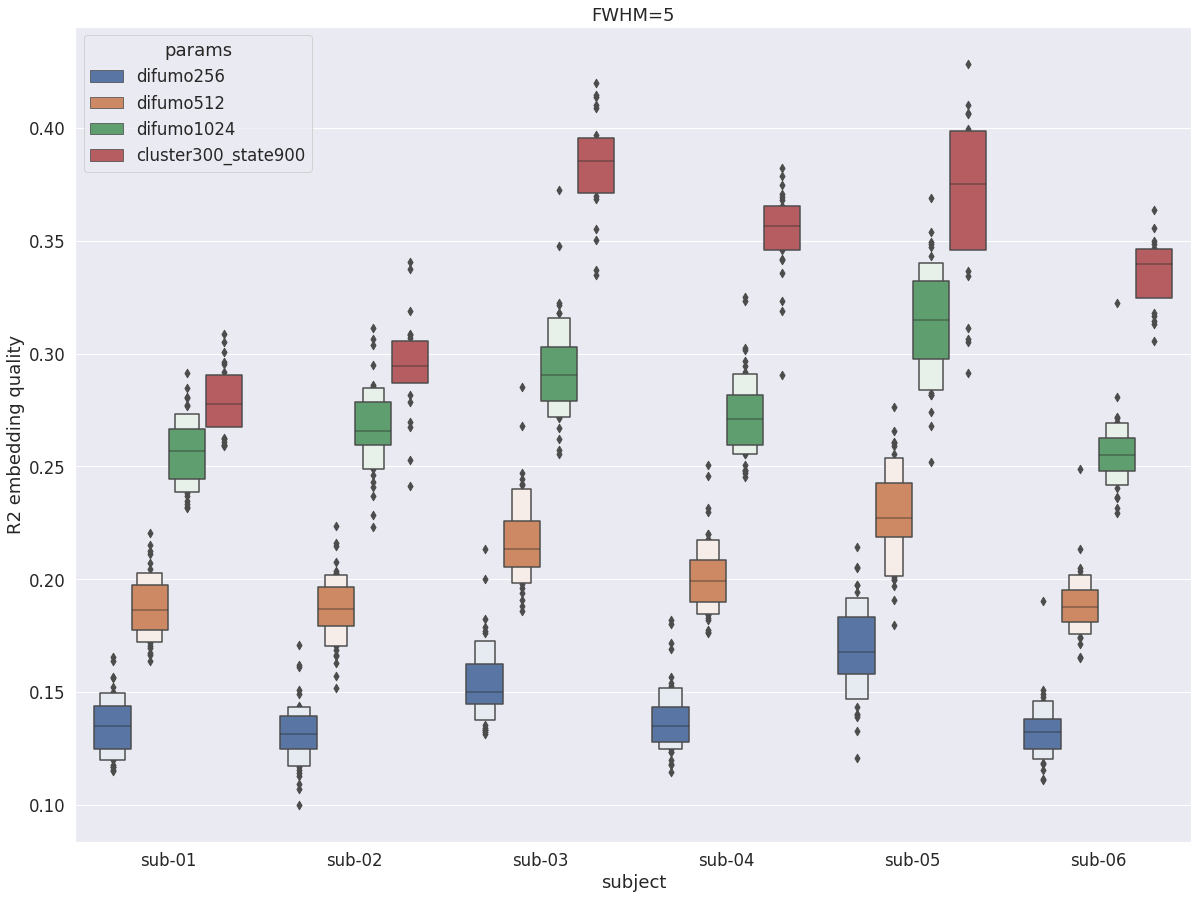
R2 with fwhm=5¶
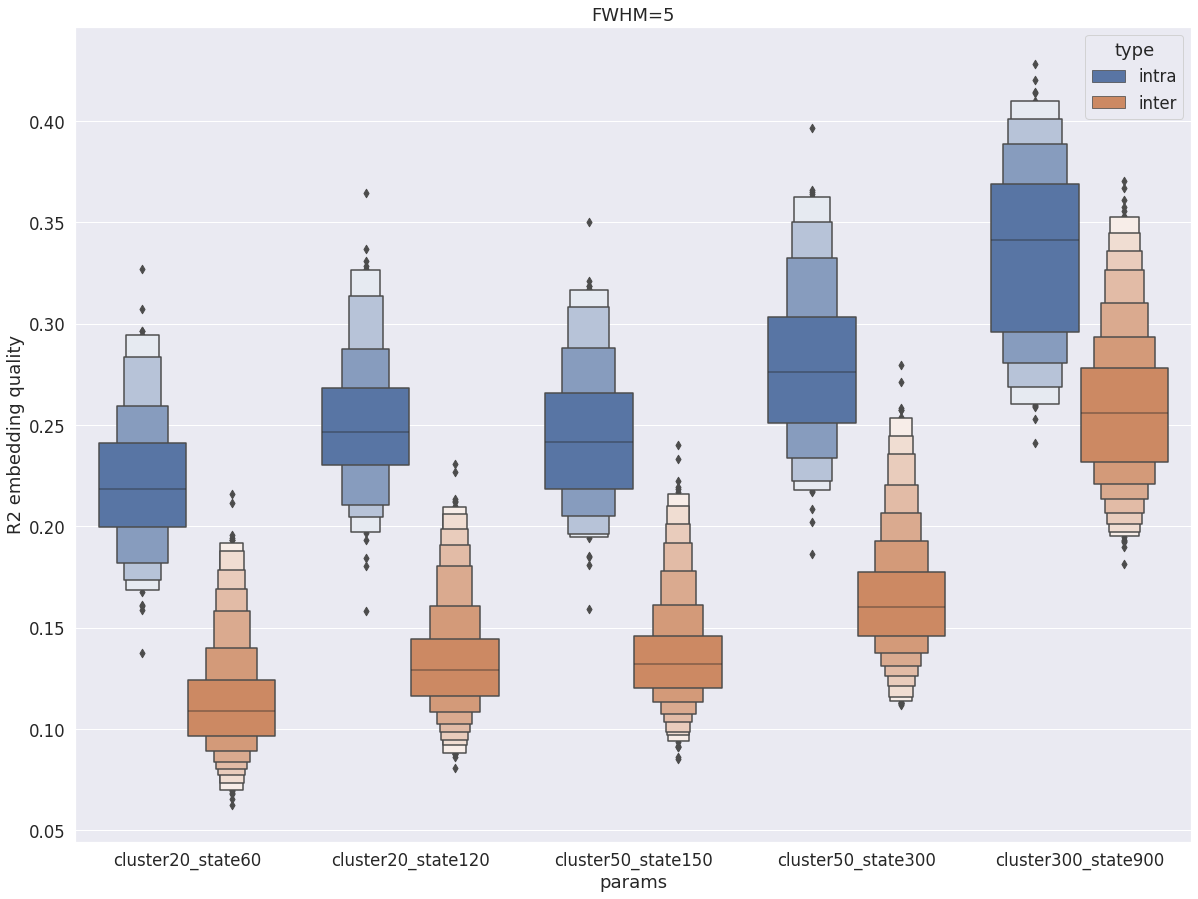
Overall, with fwhm=5, the results are very similar to what was observed in the cortex, except the R2 values are lower (by about 0.1).
DYPAC intra vs DIUFMO¶
path_data = '/home/pbellec/git/cneuromod_embeddings/cneuromod_embeddings/friends-s01/'
path_results = os.path.join(path_data, 'r2_friends-s01_cerebellum')
fwhm = 5
# DYPAC
dypac60 = os.path.join(path_results, f'r2_fwhm-intra_fwhm-{fwhm}_cluster-20_state-60.p')
dypac120 = os.path.join(path_results, f'r2_fwhm-intra_fwhm-{fwhm}_cluster-20_state-120.p')
dypac150 = os.path.join(path_results, f'r2_fwhm-intra_fwhm-{fwhm}_cluster-50_state-150.p')
dypac300 = os.path.join(path_results, f'r2_fwhm-intra_fwhm-{fwhm}_cluster-50_state-300.p')
dypac900 = os.path.join(path_results, f'r2_fwhm-intra_fwhm-{fwhm}_cluster-300_state-900.p')
# DYPAC inter-subject
inter60 = os.path.join(path_results, f'r2_fwhm-inter_fwhm-{fwhm}_cluster-20_state-60.p')
inter120 = os.path.join(path_results, f'r2_fwhm-inter_fwhm-{fwhm}_cluster-20_state-120.p')
inter150 = os.path.join(path_results, f'r2_fwhm-inter_fwhm-{fwhm}_cluster-50_state-150.p')
inter300 = os.path.join(path_results, f'r2_fwhm-inter_fwhm-{fwhm}_cluster-50_state-300.p')
inter900 = os.path.join(path_results, f'r2_fwhm-inter_fwhm-{fwhm}_cluster-300_state-900.p')
# DIFUMO
difumo256 = os.path.join(path_results, f'r2_fwhm-difumo256_fwhm-{fwhm}.p')
difumo512 = os.path.join(path_results, f'r2_fwhm-difumo512_fwhm-{fwhm}.p')
difumo1024 = os.path.join(path_results, f'r2_fwhm-difumo1024_fwhm-{fwhm}.p')
# MIST
mist197 = os.path.join(path_results, f'r2_fwhm-mist197_fwhm-{fwhm}.p')
mist444 = os.path.join(path_results, f'r2_fwhm-mist444_fwhm-{fwhm}.p')
# Schaefer
schaefer = os.path.join(path_results, f'r2_fwhm-schaefer_fwhm-{fwhm}.p')
# Smith
smith70 = os.path.join(path_results, f'r2_fwhm-smith_fwhm-{fwhm}.p')
val_r2 = pd.read_pickle(difumo256)
val_r2 = val_r2.append(pd.read_pickle(difumo512))
val_r2 = val_r2.append(pd.read_pickle(difumo1024))
val_r2 = val_r2.append(pd.read_pickle(dypac900))
fig = plt.figure(figsize=(20, 15))
sns.boxenplot(data=val_r2, x='subject', y='r2', hue='params', scale='area')
plt.ylabel('R2 embedding quality')
plt.title(f'FWHM={fwhm}')
Text(0.5, 1.0, 'FWHM=5')
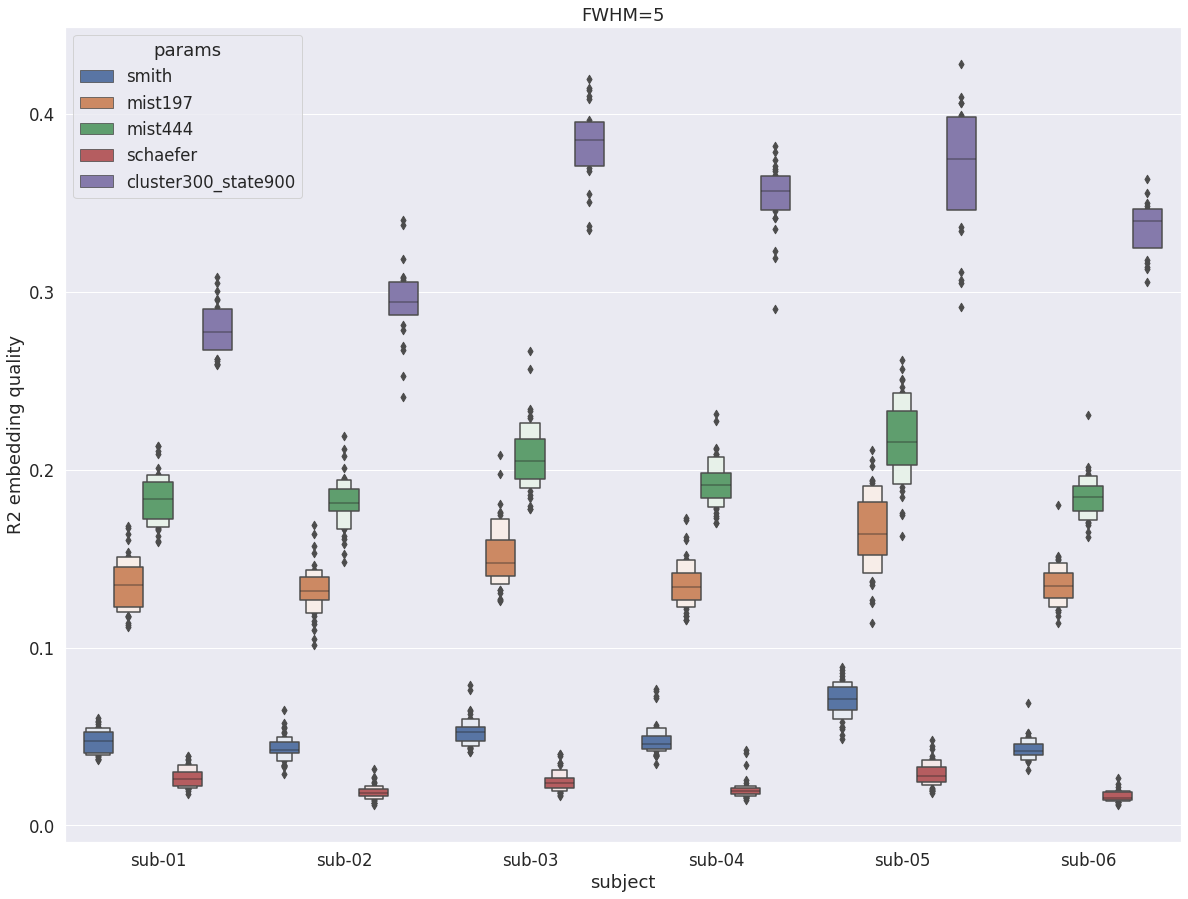
DYPAC intra vs other group parcellations¶
val_r2 = pd.read_pickle(smith70)
val_r2 = val_r2.append(pd.read_pickle(mist197))
val_r2 = val_r2.append(pd.read_pickle(mist444))
val_r2 = val_r2.append(pd.read_pickle(schaefer))
val_r2 = val_r2.append(pd.read_pickle(dypac900))
fig = plt.figure(figsize=(20, 15))
sns.boxenplot(data=val_r2, x='subject', y='r2', hue='params', scale='area')
plt.ylabel('R2 embedding quality')
plt.title(f'FWHM={fwhm}')
Text(0.5, 1.0, 'FWHM=5')
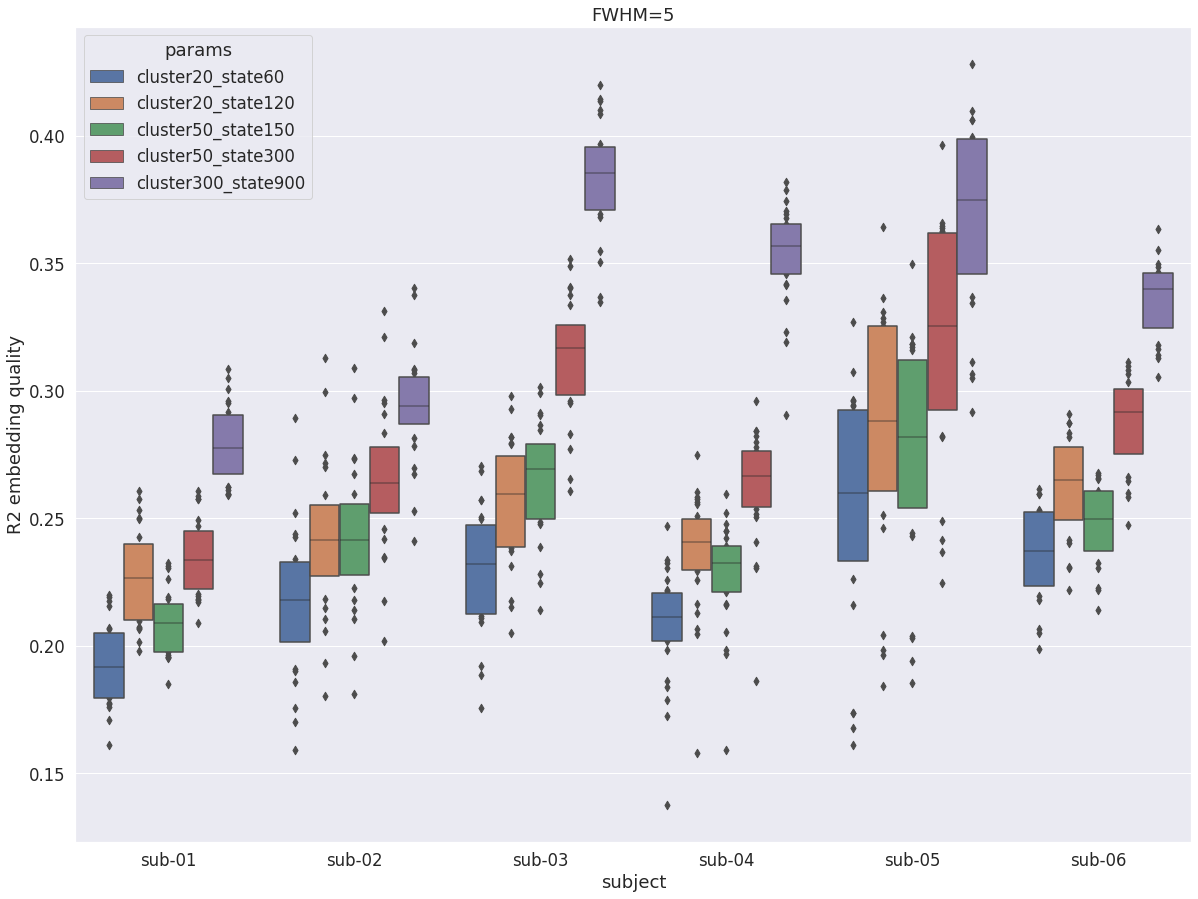
DYPAC multi-resolution¶
val_r2 = pd.read_pickle(dypac60)
val_r2 = val_r2.append(pd.read_pickle(dypac120))
val_r2 = val_r2.append(pd.read_pickle(dypac150))
val_r2 = val_r2.append(pd.read_pickle(dypac300))
val_r2 = val_r2.append(pd.read_pickle(dypac900))
fig = plt.figure(figsize=(20, 15))
sns.boxenplot(data=val_r2, x='subject', y='r2', hue='params', scale='area')
plt.ylabel('R2 embedding quality')
plt.title(f'FWHM={fwhm}')
Text(0.5, 1.0, 'FWHM=5')
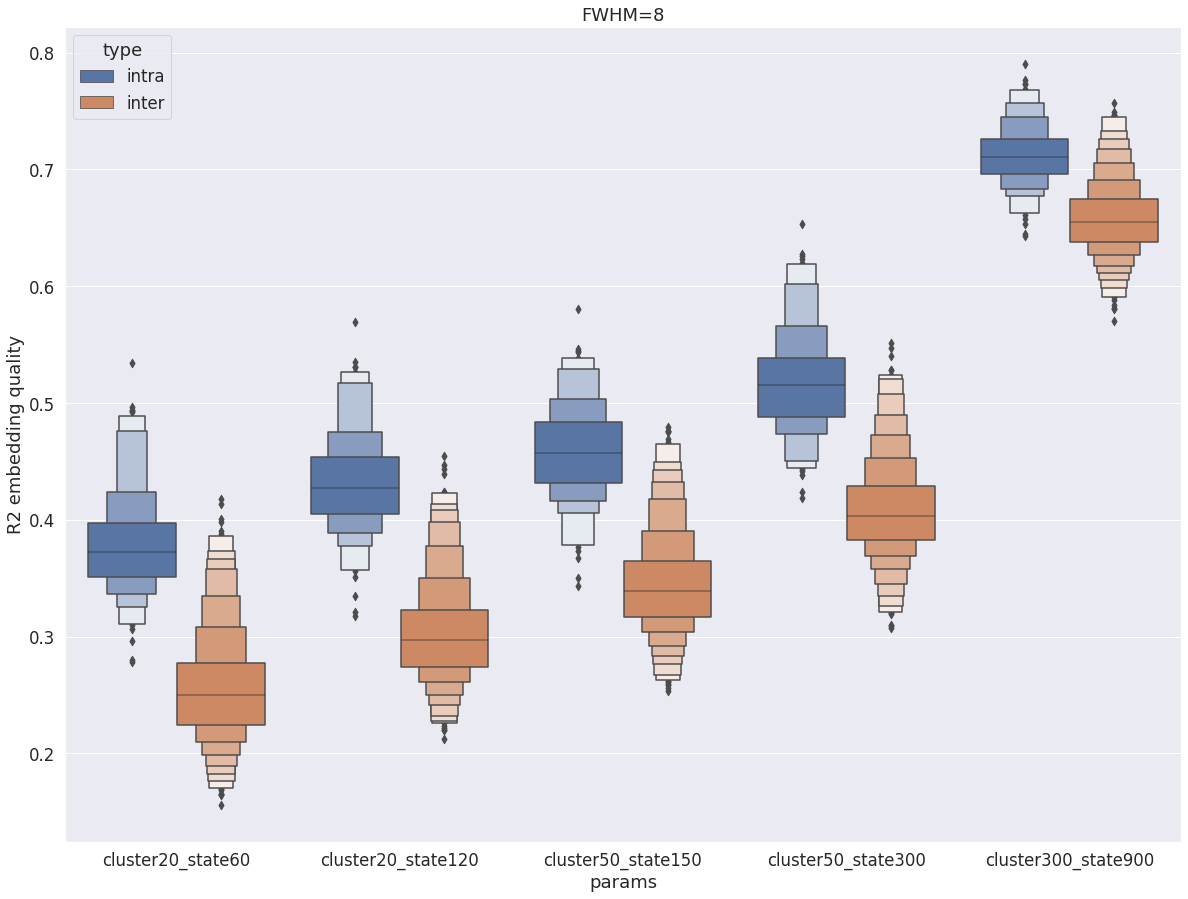
intra vs inter subject R2¶
val_r2 = pd.read_pickle(dypac60)
val_r2 = val_r2.append(pd.read_pickle(dypac120))
val_r2 = val_r2.append(pd.read_pickle(dypac150))
val_r2 = val_r2.append(pd.read_pickle(dypac300))
val_r2 = val_r2.append(pd.read_pickle(dypac900))
val_r2 = val_r2.append(pd.read_pickle(inter60))
val_r2 = val_r2.append(pd.read_pickle(inter120))
val_r2 = val_r2.append(pd.read_pickle(inter150))
val_r2 = val_r2.append(pd.read_pickle(inter300))
val_r2 = val_r2.append(pd.read_pickle(inter900))
fig = plt.figure(figsize=(20, 15))
sns.boxenplot(data=val_r2, x='params', y='r2', hue='type', scale='area')
plt.ylabel('R2 embedding quality')
plt.title(f'FWHM={fwhm}')
Text(0.5, 1.0, 'FWHM=5')
R2 with fwhm=8¶
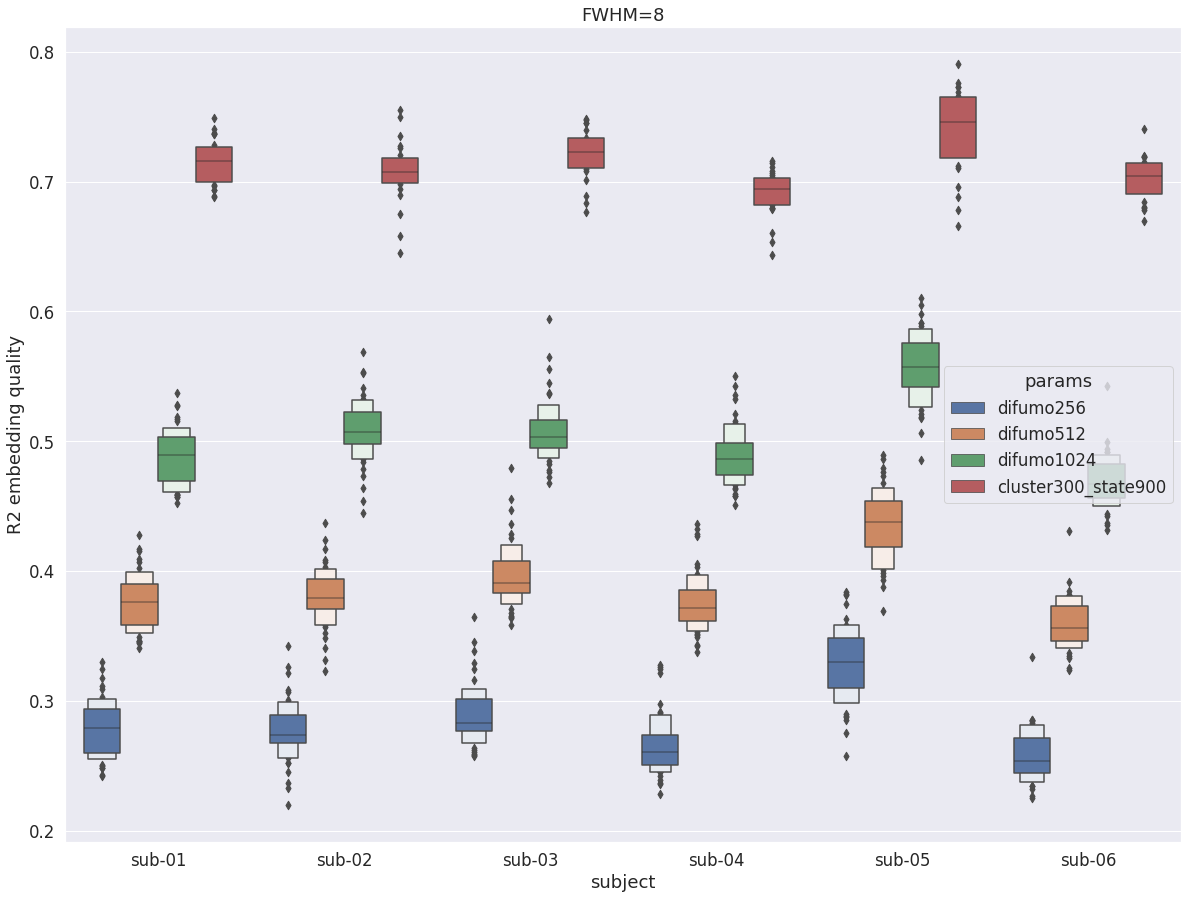
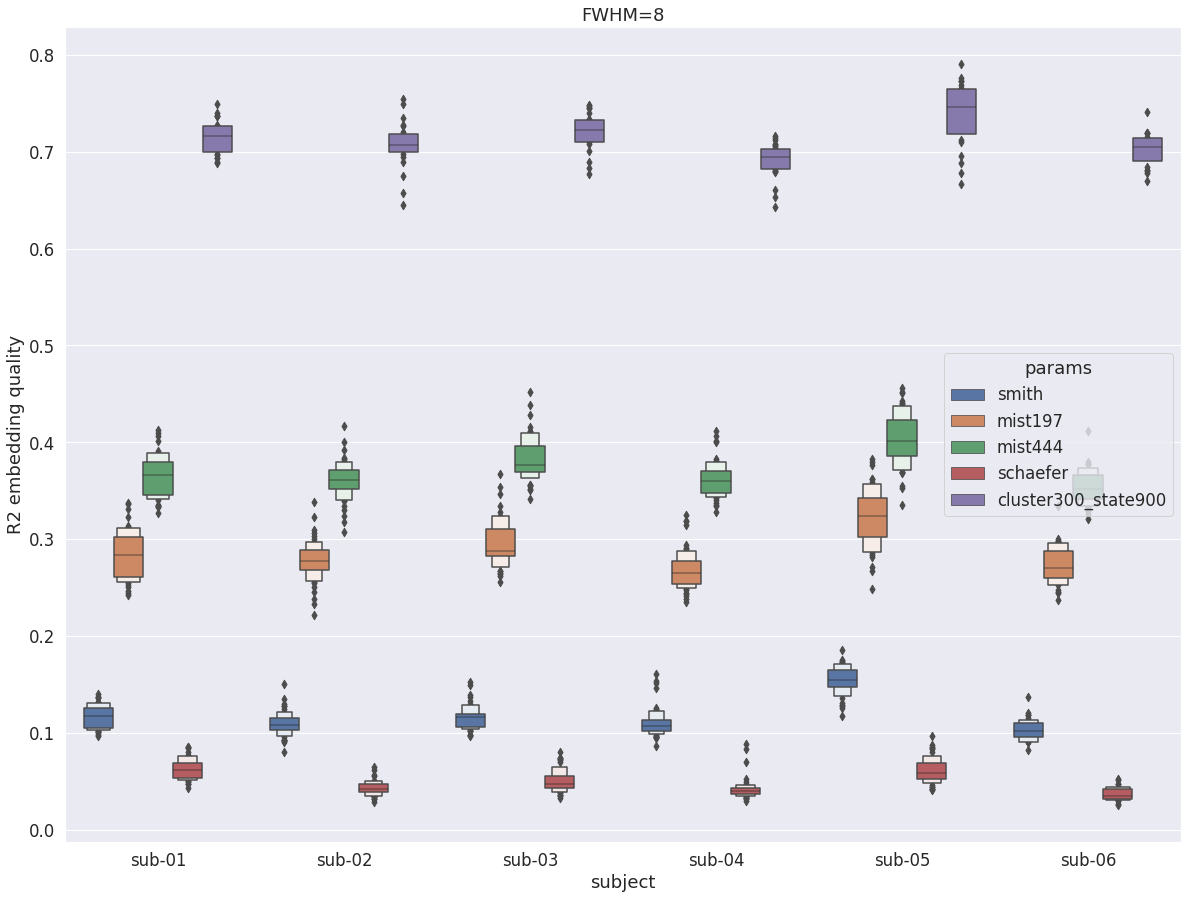
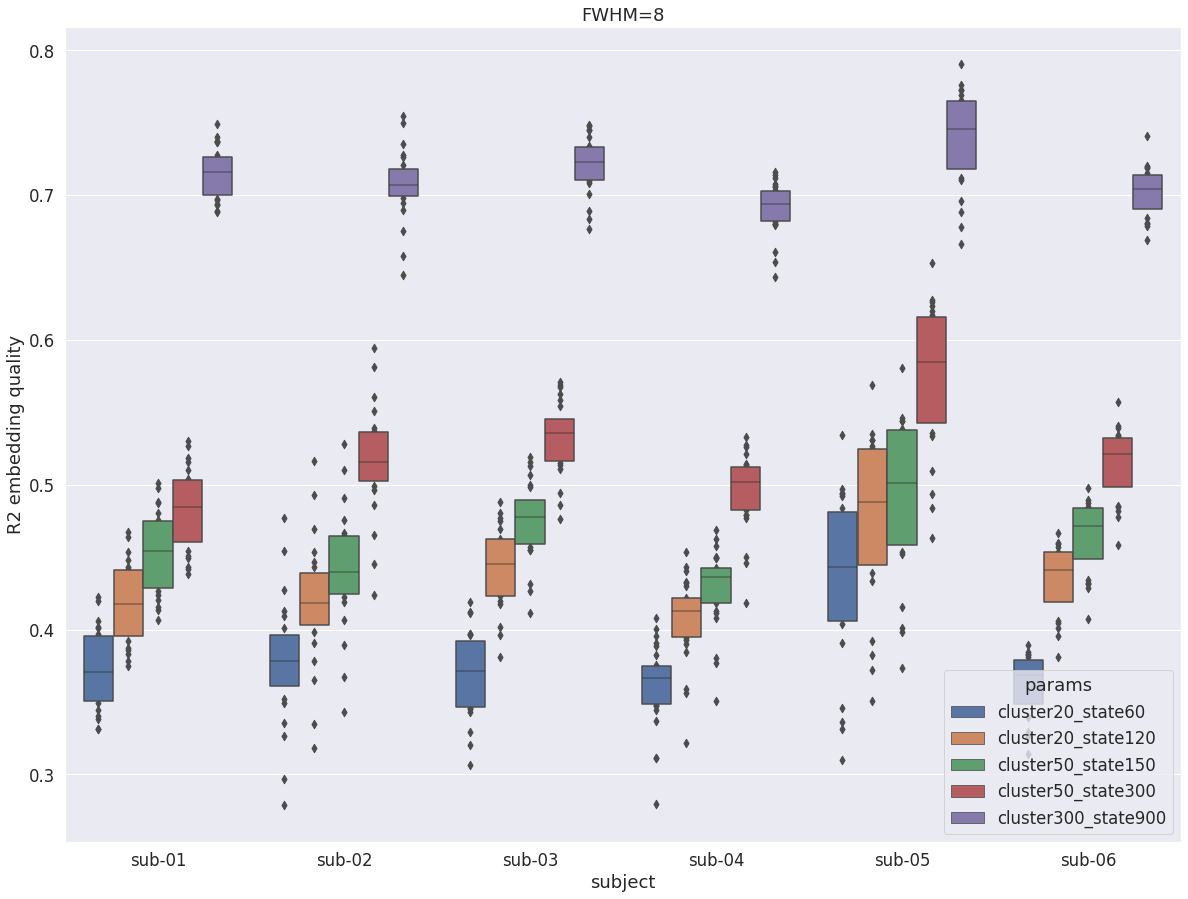
Overall, with fwhm=8, the results are very similar to what was observed in the cortex. Unlike fwhm=5, the R2 values are quite similar in the cerebellum and the cortex. The only difference is that the difference between DYPAC and DIFUMO is more pronounced in the cerebellum (i.e. DIFUMO does not perform as well in the cerebellum as in the cortex for fwhm=8), and the improvement in R2 for cluster-300_state-900 is more important in the cerebellum than in the cortex.
path_results = '/data/cisl/pbellec/cneuromod_embeddings/xp_202012/r2_friends-s01_cerebellum/'
fwhm = 8
# DYPAC
dypac60 = os.path.join(path_results, f'r2_fwhm-intra_fwhm-{fwhm}_cluster-20_state-60.p')
dypac120 = os.path.join(path_results, f'r2_fwhm-intra_fwhm-{fwhm}_cluster-20_state-120.p')
dypac150 = os.path.join(path_results, f'r2_fwhm-intra_fwhm-{fwhm}_cluster-50_state-150.p')
dypac300 = os.path.join(path_results, f'r2_fwhm-intra_fwhm-{fwhm}_cluster-50_state-300.p')
dypac900 = os.path.join(path_results, f'r2_fwhm-intra_fwhm-{fwhm}_cluster-300_state-900.p')
# DYPAC inter-subject
inter60 = os.path.join(path_results, f'r2_fwhm-inter_fwhm-{fwhm}_cluster-20_state-60.p')
inter120 = os.path.join(path_results, f'r2_fwhm-inter_fwhm-{fwhm}_cluster-20_state-120.p')
inter150 = os.path.join(path_results, f'r2_fwhm-inter_fwhm-{fwhm}_cluster-50_state-150.p')
inter300 = os.path.join(path_results, f'r2_fwhm-inter_fwhm-{fwhm}_cluster-50_state-300.p')
inter900 = os.path.join(path_results, f'r2_fwhm-inter_fwhm-{fwhm}_cluster-300_state-900.p')
# DIFUMO
difumo256 = os.path.join(path_results, f'r2_fwhm-difumo256_fwhm-{fwhm}.p')
difumo512 = os.path.join(path_results, f'r2_fwhm-difumo512_fwhm-{fwhm}.p')
difumo1024 = os.path.join(path_results, f'r2_fwhm-difumo1024_fwhm-{fwhm}.p')
# MIST
mist197 = os.path.join(path_results, f'r2_fwhm-mist197_fwhm-{fwhm}.p')
mist444 = os.path.join(path_results, f'r2_fwhm-mist444_fwhm-{fwhm}.p')
# Schaefer
schaefer = os.path.join(path_results, f'r2_fwhm-schaefer_fwhm-{fwhm}.p')
# Smith
smith70 = os.path.join(path_results, f'r2_fwhm-smith_fwhm-{fwhm}.p')
DYPAC intra vs DIFUMO¶
val_r2 = pd.read_pickle(difumo256)
val_r2 = val_r2.append(pd.read_pickle(difumo512))
val_r2 = val_r2.append(pd.read_pickle(difumo1024))
val_r2 = val_r2.append(pd.read_pickle(dypac900))
fig = plt.figure(figsize=(20, 15))
sns.boxenplot(data=val_r2, x='subject', y='r2', hue='params', scale='area')
plt.ylabel('R2 embedding quality')
plt.title(f'FWHM={fwhm}')
Text(0.5, 1.0, 'FWHM=8')
DYPAC intra vs other group parcellations¶
val_r2 = pd.read_pickle(smith70)
val_r2 = val_r2.append(pd.read_pickle(mist197))
val_r2 = val_r2.append(pd.read_pickle(mist444))
val_r2 = val_r2.append(pd.read_pickle(schaefer))
val_r2 = val_r2.append(pd.read_pickle(dypac900))
fig = plt.figure(figsize=(20, 15))
sns.boxenplot(data=val_r2, x='subject', y='r2', hue='params', scale='area')
plt.ylabel('R2 embedding quality')
plt.title(f'FWHM={fwhm}')
Text(0.5, 1.0, 'FWHM=8')
DYPAC multi-resolution¶
val_r2 = pd.read_pickle(dypac60)
val_r2 = val_r2.append(pd.read_pickle(dypac120))
val_r2 = val_r2.append(pd.read_pickle(dypac150))
val_r2 = val_r2.append(pd.read_pickle(dypac300))
val_r2 = val_r2.append(pd.read_pickle(dypac900))
fig = plt.figure(figsize=(20, 15))
sns.boxenplot(data=val_r2, x='subject', y='r2', hue='params', scale='area')
plt.ylabel('R2 embedding quality')
plt.title(f'FWHM={fwhm}')
Text(0.5, 1.0, 'FWHM=8')
Intra vs inter subject R2¶
val_r2 = pd.read_pickle(dypac60)
val_r2 = val_r2.append(pd.read_pickle(dypac120))
val_r2 = val_r2.append(pd.read_pickle(dypac150))
val_r2 = val_r2.append(pd.read_pickle(dypac300))
val_r2 = val_r2.append(pd.read_pickle(dypac900))
val_r2 = val_r2.append(pd.read_pickle(inter60))
val_r2 = val_r2.append(pd.read_pickle(inter120))
val_r2 = val_r2.append(pd.read_pickle(inter150))
val_r2 = val_r2.append(pd.read_pickle(inter300))
val_r2 = val_r2.append(pd.read_pickle(inter900))
fig = plt.figure(figsize=(20, 15))
sns.boxenplot(data=val_r2, x='params', y='r2', hue='type', scale='area')
plt.ylabel('R2 embedding quality')
plt.title(f'FWHM={fwhm}')
Text(0.5, 1.0, 'FWHM=8')